(Back | Contents
| Next)
Defining The Migration Matrix
(Note: it is recommended that you be familiar with the material
in the data file conversion
section before reading this section.)
The Migration Matrix defines how populations mix. Each cell displays the parameters for migration from one population to another. The populations may mix randomly, symmetrically, at a constant rate, or not at all. If they mix, various parameters including the rate can be defined. The values in the cells can be edited to match the user's current model. This same format is used for displaying and defining Divergence which is discussed later.
Smart modeling of how your populations mix will yield much better results from LAMARC's analysis.
Think about how your populations relate to each other geographically or temporally. For example,
if you are looking at populations in a series of valleys separated by mountains,
symmetrical migration makes sense. But if you are looking at barnicle populations
along an island chain that has a strong prevailing current, migration downstream
is easy but migration upstream isn't. You should not infer symmetrical rates here, and
perhaps should consider setting the upstream rates to zero.
Of course, if you are not sure what model to use, try an ensemble of runs testing each model and compare the results. Each will give you different insights into your data. Coalescence analysis is a tool to investigate your data not an oracle.
This is the Migration Matrix tab of the LAMARC Converter interface which displays the currently defined Migration Matrix.
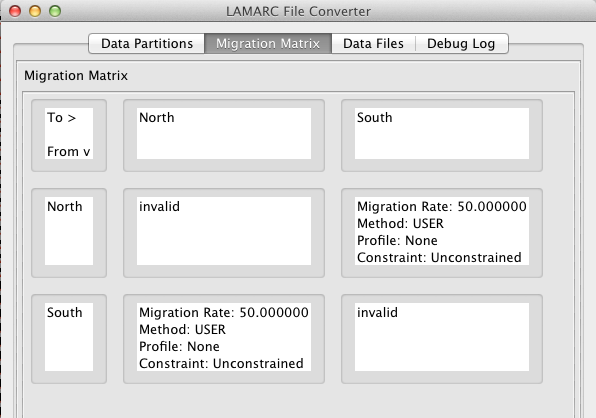
There are several things you should note:
-
The "From" Population is listed at the start of each row and the "To" Population is at the top of each column.
-
A population cannot migrate to itself, so all the cells on the diagonal are marked "invalid"
-
Only cells with black text can be edited. Those that are grayed out cannot. So if you want to modify a population name you have to go back to the Data Partitions tab.
By default LAMARC allows unconstrained migration between all populations in your data set and sets the rates all equal. If you wish to change any of the values, double click on an editable cell and the following dialog will appear:
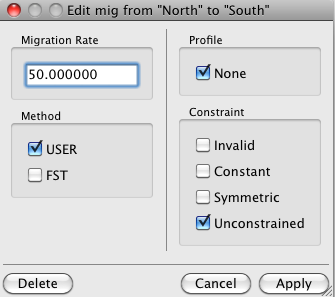
You can edit four settings here:
-
Migration Rate : Any positive real value. This represents m/(mu), the ratio of the
per-lineage migration rate to the per-site mutation rate.
-
Method : Toggle between
-
"USER" : User defined (the starting value you give will be used).
-
"FST" : Starting values will be determined using the FST statistic if possible;
otherwise your value will be used.
-
Profile : This determines whether the program will compute profiles (slices
through the posterior curve to show its shape and give approximate confidence intervals).
In a Bayesian run LAMARC will always do percentile profiles--the most informative type--
because there is minimal computational cost to doing so. In a likelihood run, however,
profiles can be quite expensive, and you may choose to ask for fixed profiles or no profiles
instead. Thus, the three options here are:
- "percentile": Percentile profiles (most expensive)
- "fixed": Fixed profiles (somewhat expensive)
- "none": No profiles (usually not recommended as you will get no information
about the confidence intervals of your estimates)
-
Constraint : Select from:
-
"Invalid" : Migration cannot happen between the "From" Population and the "To" Population.
-
"Constant" : The Migration rate will be held constant at the starting value you
give. If that value is zero, this will have the same effect as Invalid. Use this option when
you have an idea of the migration rate and do not want to infer it.
-
"Symmetric" The Migration Rate between the "From" Population and
the "To" Population must be the same as that between the "To"
Population and the "From". The effect of this it to change both
the Migration Rate and Constraint of the symmetric cell (across
the diagonal) to the values of the cell being edited. It also tells LAMARC that the
two linked rates must remain equal through out the analysis. Only one value is inferred,
and it will be the one that best fits evidence from both populations.
-
"Unconstrained" This migration rate will be inferred, starting at the given
value. This is the normal setting for a parameter of interest.
Generally you will want to make your migration rates Unconstrained unless that is
leading to inferring too many parameters, which it easily can. For cases in which
there is no reason to expect asymmetry, Symmetrical rates greatly reduce the number
of parameters. If your populations are laid out in such a way that only a few
migration routes are feasible, you should set all other routes to Invalid. Finally,
setting rates to Constant is relatively seldom used, but is useful when you already
have a Migration rate estimate in hand and want to concentrate your statistical efforts on some
other parameter.
(Back | Contents
| Next)


