SNP panel data is an increasingly popular input to sequence analysis programs. Unlike fully sequenced data, this input type is the result of a two-step process:
This process is popular because it can be used to inexpensively gather precise estimates of the rates of variation at the measured SNPs without the cost of exhaustively sequencing all study samples. While this type of data can be used to measure patterns of common variation in genetic data, it is not ideal for coalescent analysis.
Like other coalescent programs, LAMARC searches the space of coalescent trees, concentrating on those matching the input sequence data best. The SNP panel process removes most of the information available about recent variation, which is where most of the tree structure exists. LAMARC's panel correction feature compensates for this loss of data. If it is not used when it should be, estimates of Theta will be artificially low.
If your sequence data came from a SNP chip, you must use panel correction. Otherwise you will underestimate Theta, and if you turn on estimation of growth you will badly underestimate the growth rate. The strength of this bias is worst for small panels, but we do not recommend uncorrected use of panel SNP data from panels of any size.
Additional information beyond the produced sequences is required. You need to know:
This information is not always readily available. If you are using a commercial SNP chip, you may need to contact the manufacturer for this information.
If you designed and had fabricated a SNP chip created from your own panel members, you should consider performing your coalescent analysis on the original, completely sequenced set of panel members. The SNP chip will be useful for measuring allele frequencies, but searching coalescent trees is much more accurate on data that is not missing rare and low-frequency SNPs.
Additionally, you should determine if your SNP chip was constructed with a minor allele frequency cutoff, in which alleles occuring with low frequency in the panel are omitted from the chip. It is critical to know what cutoff was used.
Caution: results are highly dependent on having accurate panel size information. Therefore,
If you don't know how many panel sequences were used to create the SNP chip, Do Not Estimate The Count! You cannot perform an accurate coalescent analysis on your data.
If your panel SNPs are limited to positions which met a Minor Allele Frequency (MAF) cutoff, your panel size must be adjusted to compensate for this effect. Please email us at lamarc@u.washington.edu for help with this calculation.
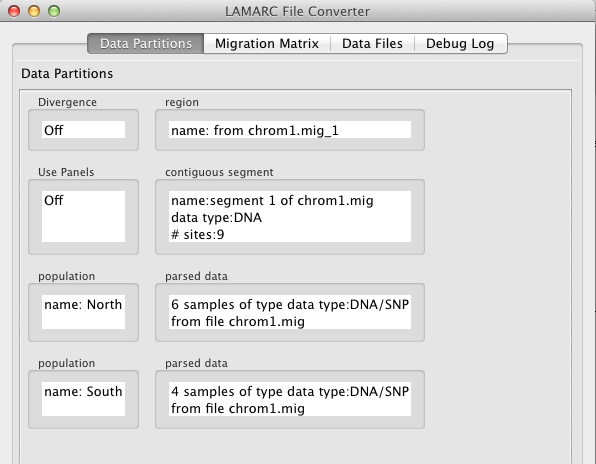
By default, panel corrections are not turned on in the converter.

To turn panels on, double click inside the box labeled "Use Panels". The initial panel size will be zero, which is equivalent to no panel correction.
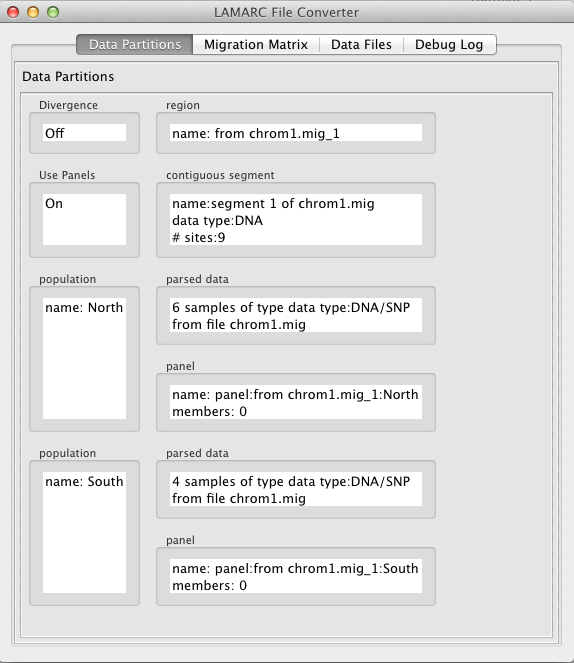
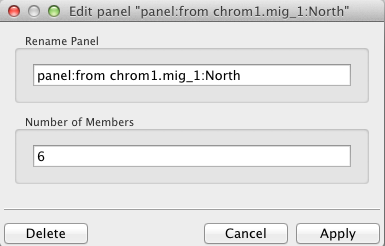
To edit the count and optionally name the panel, double click inside the box labeled "panel". It will look like this:
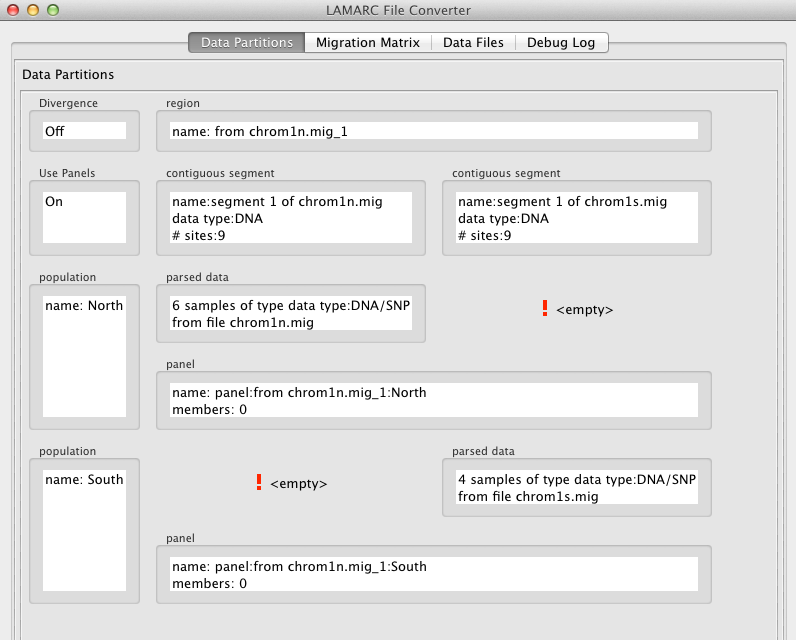
One possible source of confusion is that Panels are associated with Region / Population pairs, not with Contiguous Segments (loci close enough to each other to be in linkage disequilibrium). If you combine two or more Regions in the Converter Editor, the Loci remain separate, but they are now associated with the same Region. The other selected Regions are eliminated by the combine process. This means that Panel information associated with any Population that spaned the initial Region set must be consolidated. As there is no way to know which member count is correct, the largest value becomes the value for the combined Region. The resulting screen (created by separating the orignal input file into two separate files, reading them both in, and combining the Regions) looks like this:
The other side of this is that if you decide to separate multiple Loci that are associated with a single Region (as above), multiple new Regions are produced. The Panel member count associated with the original Region will be associated with the Panels now attached to each of the new Regions.
You can add a panel definition directly into an existing LAMARC XML file via hand-editing. See the XML Input Format documentation on panels for an example.